3D Imaging of Kidney Proximal Tubule Microphysiological Systems
Microphysiological Systems (MPS) are 3D culture systems designed to mimic in vivo microenvironments. They are gaining attention as tools for preclinical pharmaceutical evaluation, offering a more physiologically-relevant human model. They also offer an ethical alternative to animal testing. MPS utilizes components such as microfluidic channels and porous membranes to faithfully recreate the microenvironment surrounding cells in 3D. This enables research in conditions that are closer to living tissues than those possible with conventional 2D cultures. MPS is already being applied in diverse research areas, including regenerative medicine, drug development, toxicity evaluation, and disease modeling, and is expected to become a new standard for in vitro evaluation.
This application note presents the observation of primary cilia of human renal proximal tubule cells within a collagen gel-based proximal tubule model using the Nikon AX R confocal microscope system. AX R provides wider fields of view and higher 3D resolution compared to conventional confocal microscopes, enabling clear visualization of fine structures such as primary cilia (approximately 200 nm in diameter) throughout the tissue.
Keywords: microphysiological system (MPS), organ-on-a-chip, 3D culture, confocal microscopy, proximal tubule, primary cilia, regenerative medicine, drug development, toxicity evaluation, disease research
3D imaging of the proximal tubule model within the MPS chip
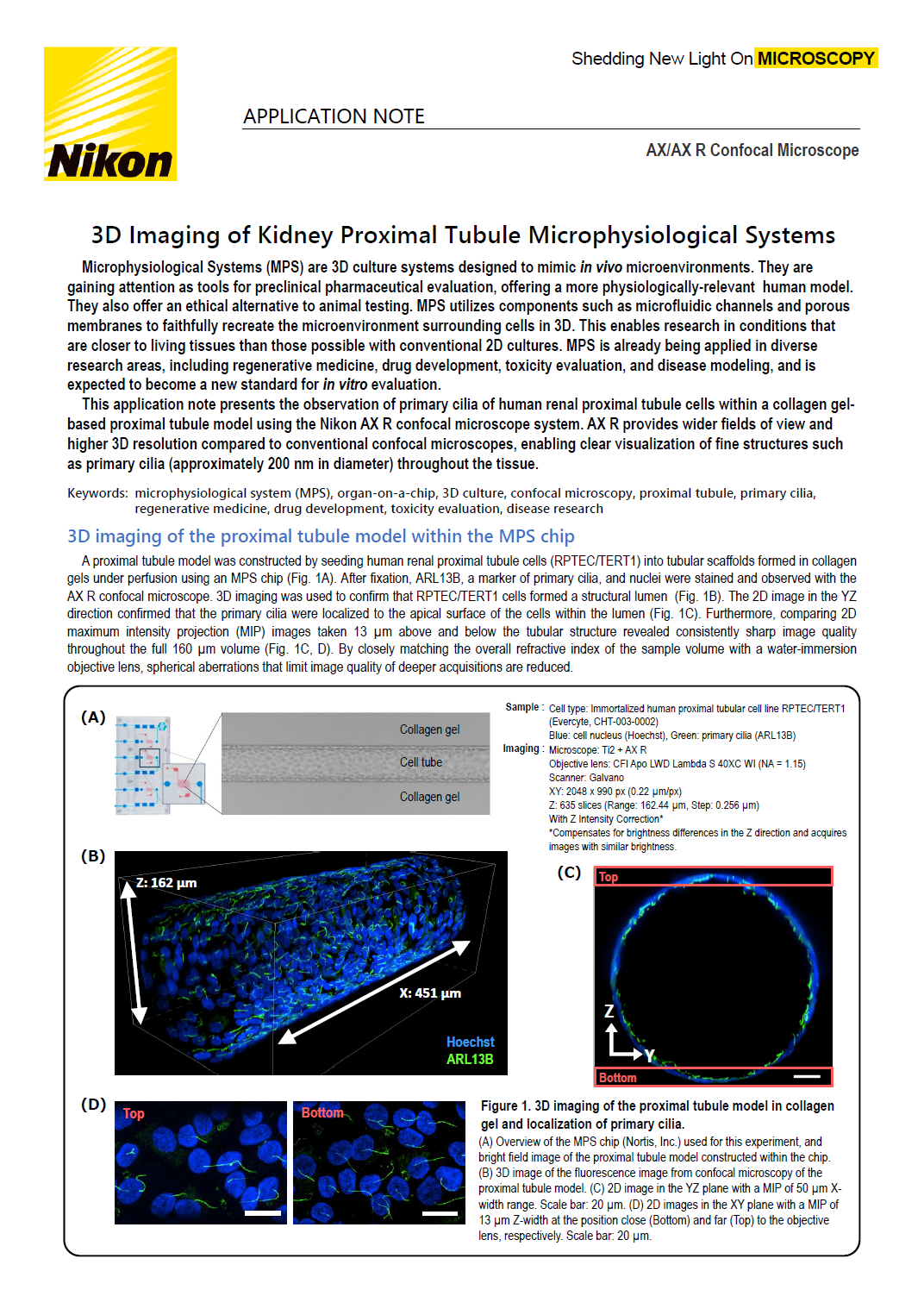
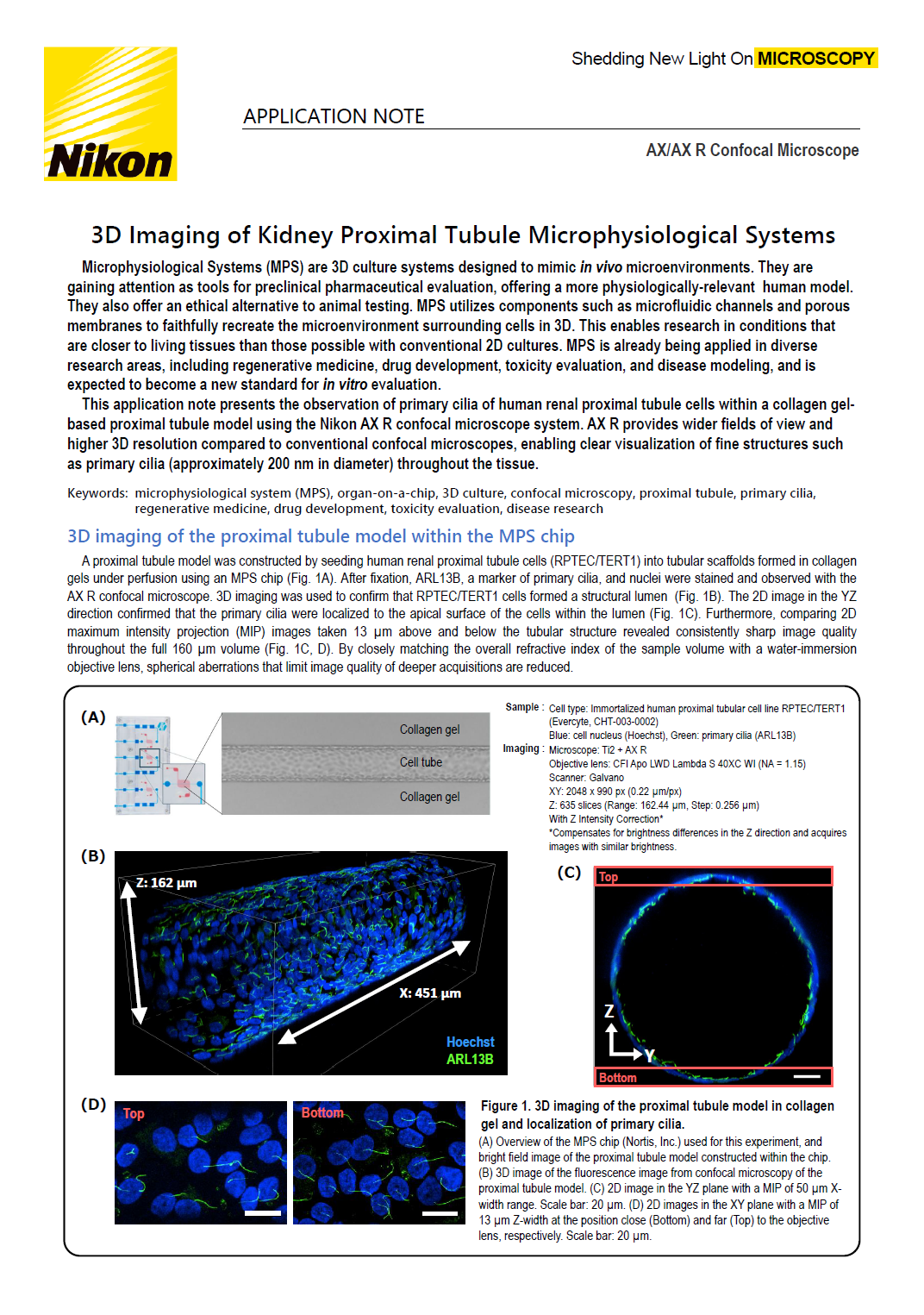
A proximal tubule model was constructed by seeding human renal proximal tubule cells (RPTEC/TERT1) into tubular scaffolds formed in collagen gels under perfusion using an MPS chip (Fig. 1A). After fixation, ARL13B, a marker of primary cilia, and nuclei were stained and observed with the AX R confocal microscope. 3D imaging was used to confirm that RPTEC/TERT1 cells formed a structural lumen (Fig. 1B). The 2D image in the YZ direction confirmed that the primary cilia were localized to the apical surface of the cells within the lumen (Fig. 1C). Furthermore, comparing 2D maximum intensity projection (MIP) images taken 13 µm above and below the tubular structure revealed consistently sharp image quality throughout the full 160 µm volume (Fig. 1C, D). By closely matching the overall refractive index of the sample volume with a water-immersion objective lens, spherical aberrations that limit image quality of deeper acquisitions are reduced.
Sample : Cell type: Immortalized human proximal tubular cell line RPTEC/TERT1
(Evercyte, CHT-003-0002)
Blue: cell nucleus (Hoechst), Green: primary cilia (ARL13B)
Imaging : Microscope: Ti2 + AX R
Objective lens: CFI Apo LWD Lambda S 40XC WI (NA = 1.15)
Scanner: Galvano
XY: 2048 x 990 px (0.22 µm/px)
Z: 635 slices (Range: 162.44 µm, Step: 0.256 µm)
With Z Intensity Correction* *Compensates for brightness differences in the Z direction and acquires images with similar brightness.
Figure 1. 3D imaging of the proximal tubule model in collagen gel and localization of primary cilia.
(A) Overview of the MPS chip (Nortis, Inc.) used for this experiment, and bright field image of the proximal tubule model constructed within the chip.
(B) 3D image of the fluorescence image from confocal microscopy of the proximal tubule model.
(C) 2D image in the YZ plane with a MIP of 50 μm X-width range. Scale bar: 20 μm.
(D) 2D images in the XY plane with a MIP of 13 μm Z-width at the position close (Bottom) and far (Top) to the objective lens, respectively. Scale bar: 20 μm.
Analysis of primary cilia orientation using General Analysis 3
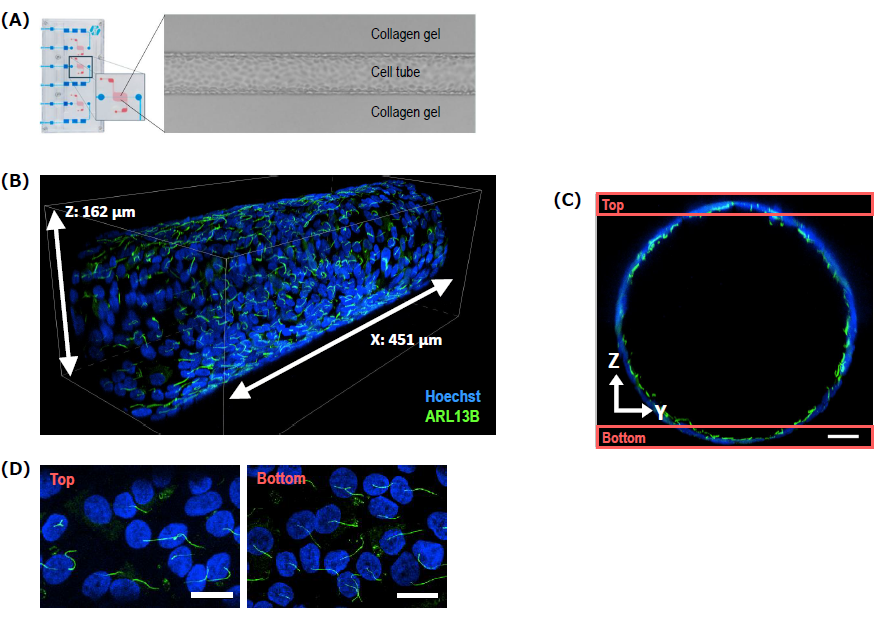
The General Analysis 3 (GA3), an an image processing and measurement module within the NIS-Elements software platform, enables customizable image analyses through a graphical user interface (GUI). We analyzed the orientation of primary cilia using GA3. We extracted the area of primary cilia in the MIP image at the bottom. For each cilium, we determined the angle corresponding to the maximum Feret's diameter* relative to the direction of media flow (X-axis) and used this value as the orientation of primary cilium (Fig. 2A, B, and C). This analysis quantitatively demonstrates that the primary cilia in the image were oriented along the direction of perfusion (Fig. 2D). GA3 provides a comprehensive suite of image analysis tools for quantifying diverse features, such as object angles and their population-level statistics (Fig. 2E).
*Feret's diameter is the distance between two parallel lines tangent to the object's boundaries in a specific direction.
Figure 2. Analysis of primary cilia orientation
(A) Image used for analysis. Scale bar: 100 μm. (B) Primary cilia (magenta) segmented using GA3. Magnified view of the region indicated by the red box in (A). Scale bar: 20 μm. (C) Calculation method of primary cilia orientation. (D) Circular histogram showing the orientation of primary cilia calculated from the image (n=85). (E) Partial view of the GA3 user interface. The "Orientation" angle measurement function was used in this analysis.
Conclusion
Utilizing the AX R confocal microscope, we captured detailed images of the proximal tubule model constructed with an MPS chip. This enabled us to clearly capture the tubular structure of RPTEC/TERT1 formed in collagen gel, as well as the structure and orientation of primary cilia along the direction of perfusion throughout the entire luminal structure.
The AX/AX R confocal microscope, combined with the NIS-Elements image analysis software, provides superior high-resolution imaging capabilities and enables detailed quantitative analysis. Employing this powerful imaging platform with MPS models allows for the acquisition of more accurate data for investigating disease mechanisms and developing novel therapies. This combined approach is expected to serve as a powerful research tool in medical and biological research, as well as in pharmaceutical development.
Authors: Takehiro Nakamura and Shingo Nagawa, Nikon Corporation
References
S Ishida, T Kanamori. Microphysiological system as a promising technology for drug assay, Folia Pharmacol. Jpn., 154, 345-351, (2019).
Anna Tourovskaia et al. Brief Communication: Tissue-engineered Microenvironment Systems for Modeling Human Vasculature, Exp. Biol. Med., 239(9): 1264–1271, (2014).
Elijah J. Weber et al. Development of a microphysiological model of human kidney proximal tubule function, Kidney Int., 90(3): 627–637, (2016).
Footnote
Primary cilia
Primary cilia are hair-like structures several μm long and approximately 200 nm in diameter that protrude from the cell surface and are important cell organelles involved in various signal transductions.
Primary cilia in the tubules sense urine flow and are involved in kidney organogenesis and maintenance. Abnormalities of these primary cilia are known to cause various renal diseases and have been studied in terms of disease mechanisms and kidney morphogenesis.
Product Information
AX/AX R Confocal Microscope
Supports high-speed, high-resolution, wide-FOV confocal imaging with low phototoxicity to living cells and reduced photobleaching.
High Speed: Up to 720 frames per second (Resonant 2048 x 16 pixels)
High resolution: Up to 8K (Galvano) / 2K (Resonant)
High throughput: Ultra-wide field of view of 25 mm
스펙