Quantitative Live-Cell Imaging to Analyze Immune Cell Secretion Activity and Gene Expression
Single-cell RNA sequencing (scRNA-seq) has revolutionized our understanding of cellular heterogeneity by identifying distinct cell types and states at the molecular level. However, its destructive nature limits insights into dynamic cellular processes. While conventional live-cell imaging can visualize cell morphology, motility, and the dynamics of a few pre- labeled molecules, it has limitations in capturing the dynamic functions of intact cells.
To overcome these limitations, researchers led by Dr. Yoshitaka Shirasaki (Research Center for Advanced Science and Technology, The University of Tokyo) and Dr. Mai Yamagishi (Live Cell Diagnosis, Ltd.) developed Live Cell Imaging of Secretion Activity (LCI-S), which combines Total Internal Reflection Fluorescence Microscopy (TIRFM) with FluoroSpot technology to enable real-time visualization and quantification of secreted proteins at the single-cell level. They further developed Time-Dependent Cell-State Selection (TDCSS), enabling selective isolation of cells in specific secretory states detected by LCI-S for subsequent scRNA-seq analysis. Using the Nikon Perfect Focus System (PFS), they achieved stable long-term imaging over five days, allowing them to capture rare cellular events and identify previously uncharacterized gene expression profiles associated with immune cell activation. This application note highlights their findings on the dynamic secretory activities and rapid gene expression changes upon activation in immune cells achieved with LCI-S and TDCSS. The collaborative research effort involves the laboratories of Dr. Shirasaki and Dr. Yamagishi, Dr. Kazuyo Moro from Department of Microbiology and Immunology, Graduate School of Medicine, Osaka University, Dr. Koichi Fukunaga from Division of Pulmonary Medicine, Department of Medicine, Keio University School of Medicine and Dr. Sotaro Uemura from Department of Biological Sciences, Graduate School of Science, The University of Tokyo.
Keywords: Live-cell imaging, Dynamic analysis, Secretory activity, Single-cell RNA sequencing, Immune cells
Visualizing Cytokine Secretion Dynamics with LCI-S
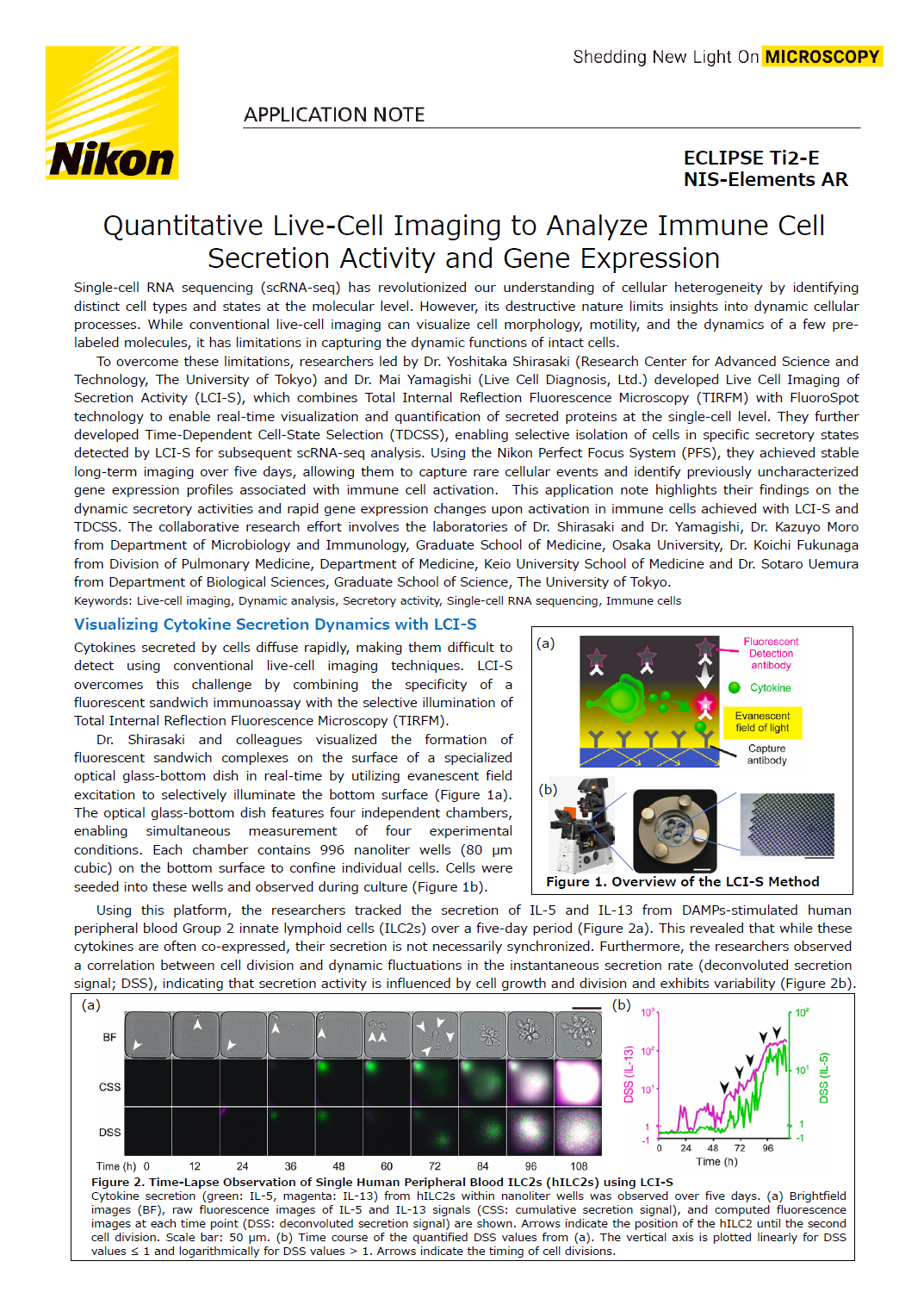
Cytokines secreted by cells diffuse rapidly, making them difficult to detect using conventional live-cell imaging techniques. LCI-S overcomes this challenge by combining the specificity of a fluorescent sandwich immunoassay with the selective illumination of Total Internal Reflection Fluorescence Microscopy (TIRFM).
Dr. Shirasaki and colleagues visualized the formation of fluorescent sandwich complexes on the surface of a specialized optical glass-bottom dish in real-time by utilizing evanescent field excitation to selectively illuminate the bottom surface (Figure 1A). The optical glass-bottom dish features four independent chambers, enabling simultaneous measurement of four experimental conditions. Each chamber contains 996 nanoliter wells (80 μm cubic) on the bottom surface to confine individual cells. Cells were seeded into these wells and observed during culture (Figure 1B).
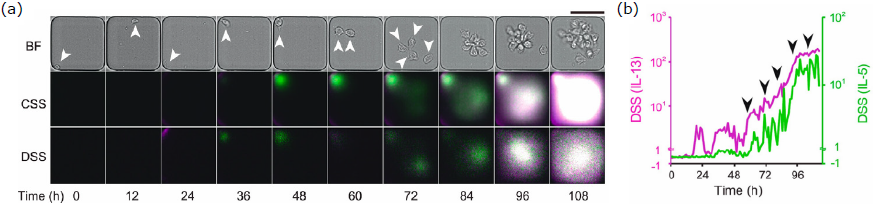
Using this platform, the researchers tracked the secretion of IL-5 and IL-13 from DAMPS-stimulated human peripheral blood Group 2 innate lymphoid cells (ILC2s) over a five-day period (Figure 2a). This revealed that while these cytokines are often co-expressed, their secretion is not necessarily synchronized. Furthermore, the researchers observed a correlation between cell division and dynamic fluctuations in the instantaneous secretion rate (deconvoluted secretion signal); DSS, indicating that secretion activity is influenced by cell growth and division and exhibits variability (Figure 2b).
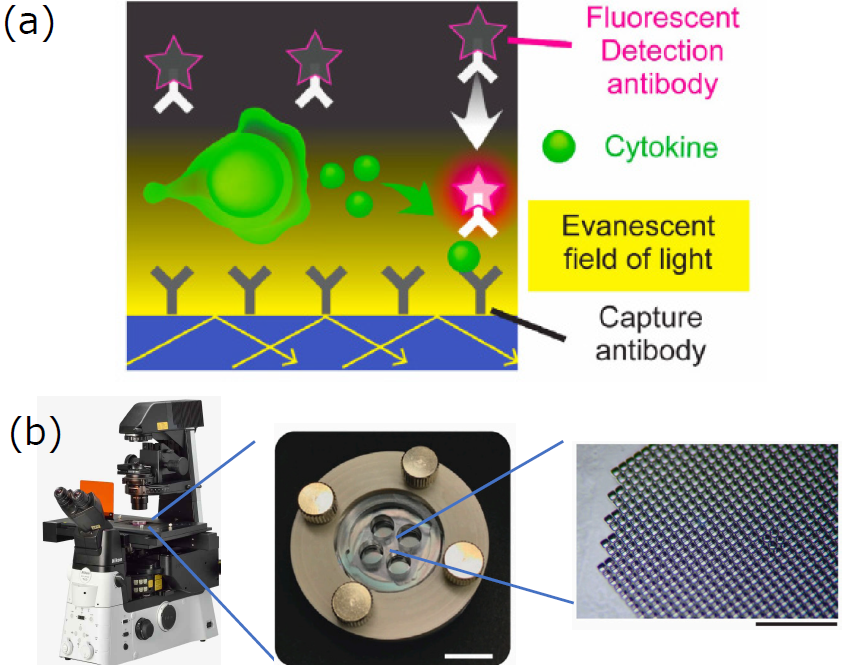
Figure 1. Overview the LCI-S Method
Figure 2. Time-Lapse Observation of Single Human Peripheral Blood ILC2s (hILC2s) using LCI-S
Cytokine secretion (green: IL-5, magenta: IL-13) from hILC2s within nanoliter wells was observed over five days. (a) Brightfield images (BF), raw fluorescence images of IL-5 and IL-13 signals (CSS: cumulative secretion signal), and computed fluorescence images at each time point (DSS: deconvoluted secretion signal) are shown. Arrows indicate the position of the hILC2 until the second cell division. Scale bar: 50 μm. (b) Time course of the quantified DSS values from (a). The vertical axis is plotted linearly for DSS values ≤ 1 and logarithmically for DSS values > 1. Arrows indicate the timing of cell divisions.
Single-Cell analysis using TDCSS
Cells exposed to stimuli are thought to transition through transient states before reaching an activated state (Figure 3a). LCI-S revealed that even when cells of the same type are exposed to the same stimulus under identical conditions, the timing of dynamic state changes (activation) accompanied by secretory functions varies (Figure 3b). Conventional scRNA-seq analysis involves "snapshot" collection of cells at a specific time point after stimulation for gene expression analysis (Figure 3c, black dashed line). This makes it difficult to capture the gene expression profiles of rare cells during transient transitional states (Figure 3d). In contrast, TDCSS offers the advantage of selectively collecting cells in specific secretory activity states, as identified through live-cell imaging with LCI-S (Figure 3c, green dashed line). By subjecting these collected cells to scRNA-seq analysis, the gene expression profiles of cells in transitional states can be directly revealed (Figure 3e).
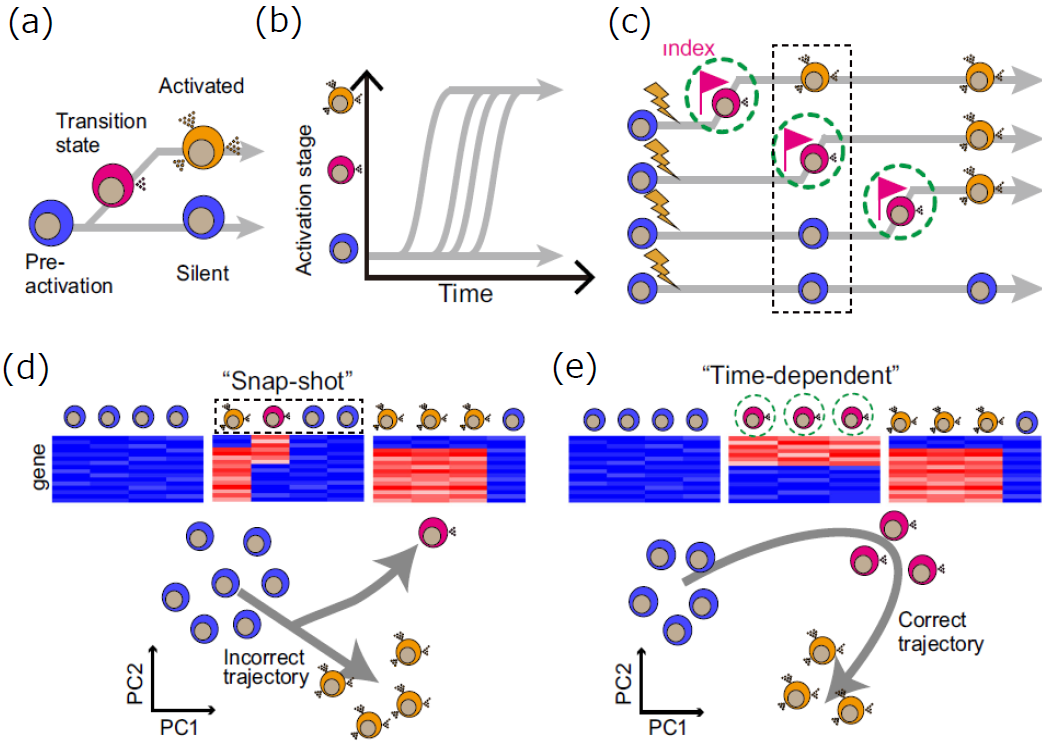
Figure 3. The concept of TDCSS
TDCSS analysis of hILC2s
hILC2s are rare cells, present at only 50-150 cells/mL in peripheral blood, and their response timing varies significantly between individual cells. Therefore, in a snapshot sample, the frequency of activating cells is extremely low, making it challenging to obtain a representative gene expression profile. To address this, two cell collection strategies were compared: 1) snapshot collection 1-2 hours after stimulation, and 2) precise collection of activating cells at defined time intervals (Δt = 30 or 60 minutes) after secretion onset (index), as determined by monitoring IL-13 secretion activity in stimulated cells using LCI-S (Figure 4a). Analysis of IL13 gene expression revealed substantial heterogeneity in the snapshot samples, whereas cells collected at defined Δt values showed significantly less variation, with greater homogeneity observed at smaller Δt values (Figure 4b). Subsequently, scRNA-seq analysis was performed on three snapshot populations (pre-DAMPS exposure (P), long after DAMPS exposure with active secretion (A), and long after DAMPS exposure without secretion (Silent)), and on activating cells collected within a defined timeframe following secretion onset (Figure 4c). This analysis revealed distinct gene expression profiles between the T and the A (Figure 4d). Based on these expression profiles, gene sets were classified as follows:
Early Induced/Reduced Genes : Gene sets induced (or reduced) immediately after the onset of secretory response
Late Induced/Reduced Genes : Gene sets with time-delayed induction (or repression) after the onset of secretory response
Transiently Induced/Reduced Genes : Gene sets induced (or reduced) only during the initial phase of secretory response
Figure 4. Single-Cell Analysis of Human Peripheral Blood ILC2s (hILC2s) using TDCSS
(a) Overview of TDCSS. Secretory activity was measured based on brightfield and fluorescence images of hILC2 cells, and cells in the transient activating state were collected. Δt represents the time elapsed between the onset of secretion and cell collection.
(b) IL13 expression levels and variability. IL13 expression levels were plotted for cells collected at Δt = 0.5 and 1 hour, and for cells collected via a snapshot approach.
(c) Fluorescence and brightfield images of hILC2s obtained by LCI-S. Representative images are shown for cells pre-stimulation (P), in the transient activating state (T), in the fully activated state (A), and in the non-secreting state long after stimulation (Silent).
(d) Gene expression profiles of the three cell states (P, T, and A). Transiently induced (repressed) gene sets are indicated as TI(R)Gs, early induced gene sets as EIGs, and late induced (repressed) gene sets as LI(R)Gs.
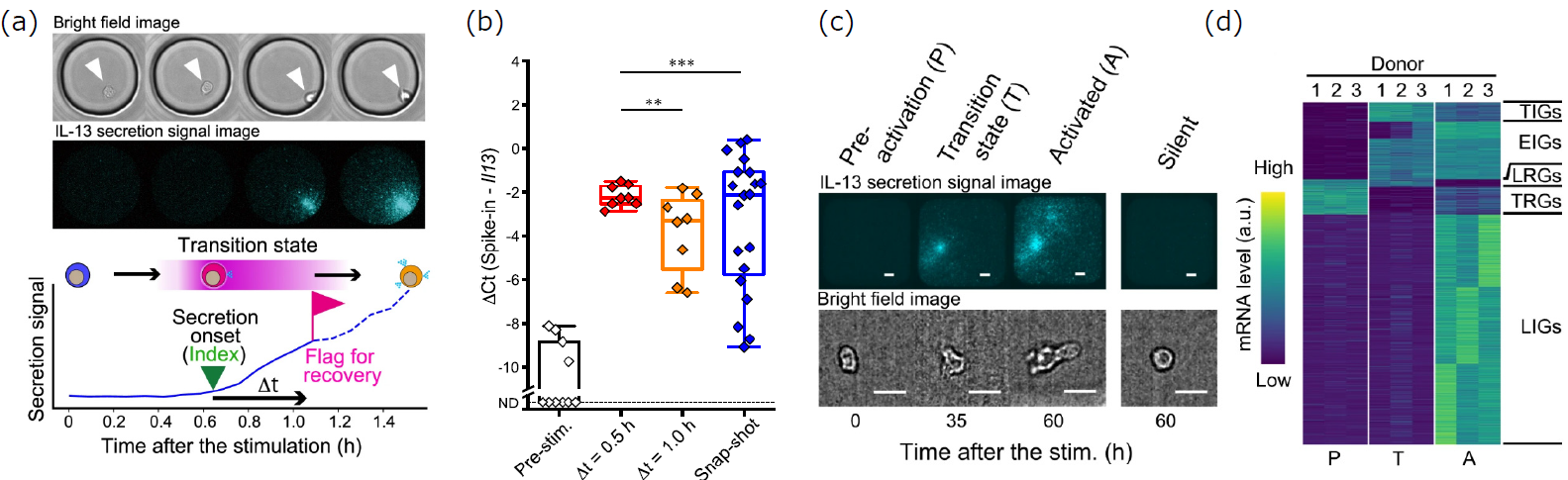
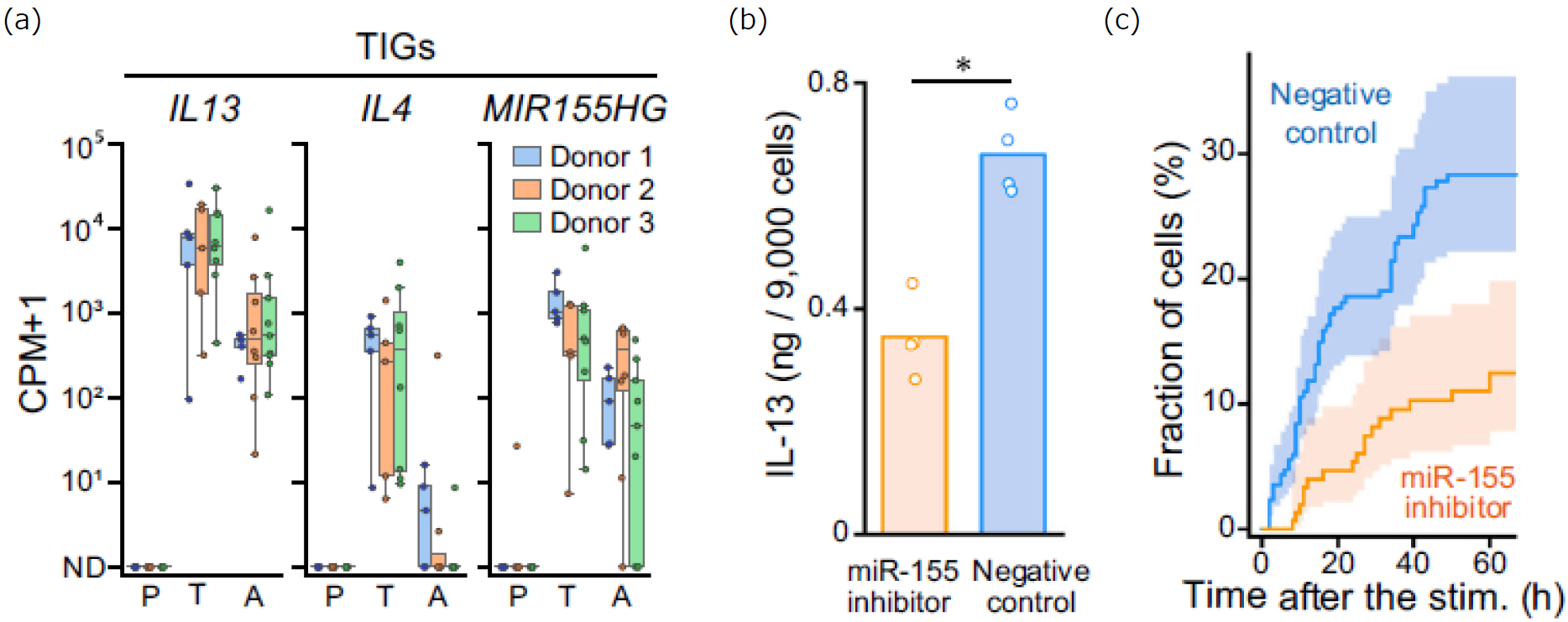
Figure 5. Analysis of a Transiently Induced Gene miR-155
(a) Inter-donor comparison of representative transiently induced genes (IL13, IL4, MIR155HG (host gene of miR-155)). Expression levels in individual cells (circles) and their distribution are shown for each activation state (P, T, and A). (b) Comparison of IL-13 production in culture supernatants with and without the miR-155 inhibitor. (c) Comparison of cumulative IL-13-secreting cell counts with and without the miR-155 inhibitor. Nelson-Aalen estimators were used. Shaded bands represent the 95% confidence intervals.
Analysis of miR-155, a Transiently Induced Gene Identified by TDCSS
The transiently induced gene miR-155 (Figure 5a) is known to be a miRNA that acts as a negative regulator of genes suppressing IL-13 secretion in hILC2s. It was hypothesized that the induction of miR-155 expression could enhance IL-13 secretion during the early phase of the secretory response. To test this, miR-155 function was inhibited using RNA interference, which resulted in a significant decrease in the number of IL-13-secreting cells (Figures 5b and 5c). Thus, TDCSS enabled the first observation of transient miR-155 expression in hILC2s, demonstrating its utility in elucidating the regulatory mechanisms of secretory responses.
Conclusions
This application note highlighted the use of LCI-S and TDCSS for analyzing the secretory activity and gene expression of rare cells. LCI-S provides a powerful method for understanding the dynamic functions of immune cells by capturing their secretory activities in real-time with high sensitivity. Combining TDCSS with scRNA-seq enables the analysis of gene expression profiles in cells at specific transitional states by allowing for the time-dependent collection of cells based on their secretory status. These techniques are facilitated by the Perfect Focus System (PFS) and multi-point time-lapse imaging capabilities of Nikon microscopes.
The NIS-Elements imaging software, also utilized in this research, offers advanced morphological analysis functions, making it a versatile single-cell analysis tool that can simultaneously acquire information on secretory activity, gene expression, and cell morphology. This integrated approach promises to provide new insights for drug discovery and life science research.
References
1. Quantitative live-cell imaging of secretion activity reveals dynamic immune responses.
Yamagishi, M., Miyata, K., Kamatani, T. et al., iScience, 27(6), 2024
( https://doi.org/10.1016/j.isci.2024.109840 )
2. Time-dependent cell-state selection identifies transiently expressed genes regulating ILC2 activation.
Tanaka, Y., Yamagishi, M., Motomura, Y. et al., Commun Biol 6, 915, 2023
( https://doi.org/10.1038/s42003-023-05297-w )
Figures in this application notes are adapted and modified with permission from authors of publications above, under the terms of the Creative Commons Attribution License (CC BY, https://creativecommons.org/licenses/by/4.0/).
Acknowledgement
We would like to express our sincere gratitude to Dr. Yoshitaka Shirasaki of the Research Center for Advanced Science and Technology, The University of Tokyo and Dr. Mai Yamagishi of the Live Cell Diagnosis, Ltd. for their great cooperation and generous advice.
Website of the Live Cell Diagnosis, Ltd. : [QR code]
https://lcd.co.jp/
Edited by Shingo Nagawa, Nikon Corporation
Product Information
ECLIPSE Ti2-E
The ECLIPSE Ti2-E's high-speed coordination with a wide variety of accessories enables efficient acquisition of reliable data, even in complex, multidimensional imaging experiments. Furthermore, its exceptional stability ensures precise focus maintenance during long-term, high-resolution observations.
스펙